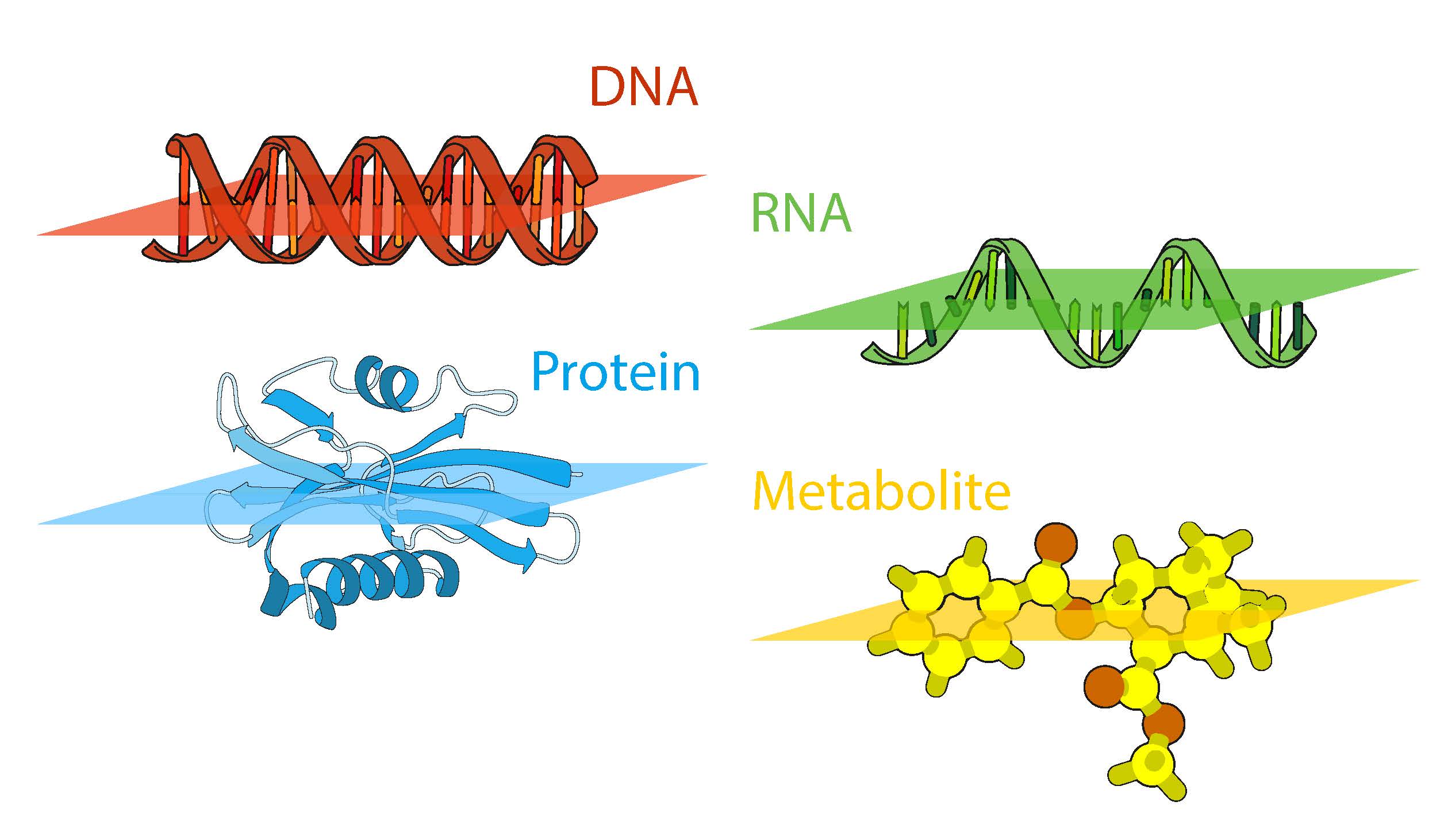
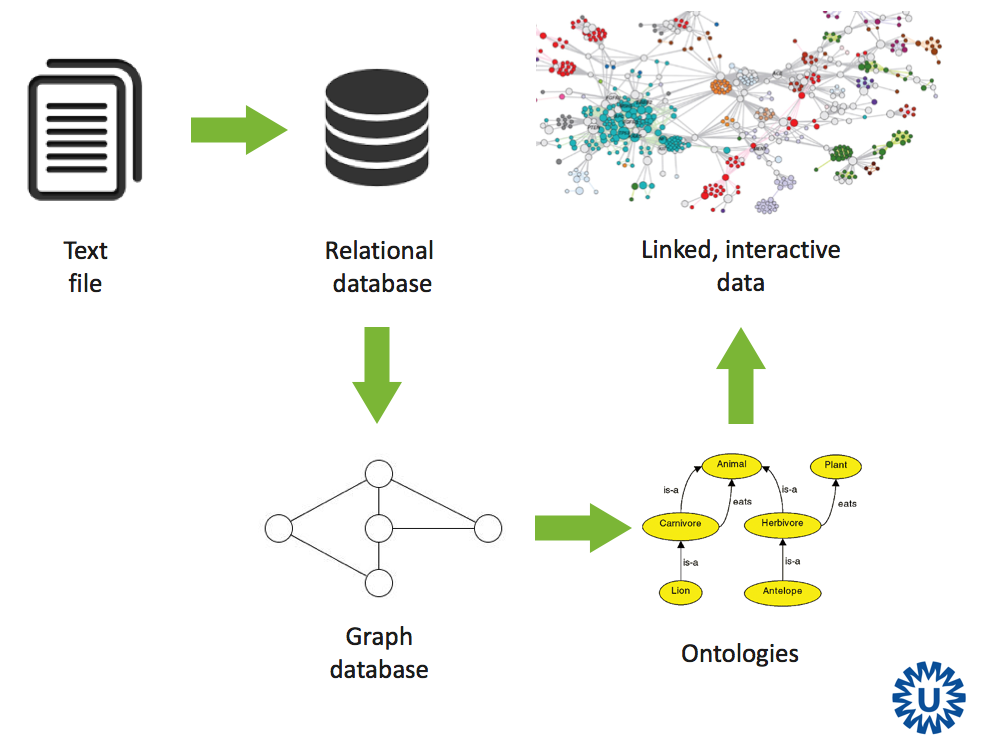
Effective mining of data and integrating data is one of the major challenges in biomedical research. Decennia of research have led to an accumulation of databases world-wide, including important resources, such as NCBI, KEGG, ENCODE, SWISS-PROT etc. Lately, new data acquisition technologies, especially next generation sequencing (NGS), are rapidly increasing the amount of information available online, from data published with papers all the way to large scale collaborations, such as The Genome Cancer Atlas (TCGA) involving a wide range of hospitals and research groups offering information on patients, diagnostics, treatments together with data on sequenced tumors, gene expression, methylation, etc. For an inspiring example see
http://www.cbioportal.org/public-portal/tumormap.do?case_id=TCGA-A2-A0CX&cancer_study_id=brca_tcga_pub.
The challenge is to effectively mine resources, such as the TCGA, after performing an experiment or getting clinical results. For example, if you are sequencing cancer tumors of patients, the question is: how to mine this public data and compare the results against your own data and results. TCGA alone numbers over 50,000 files, there is no way to mine this data by hand. Likewise we have access to 1,000 public genomes and the genome of the Netherlands (GoNL). What are feasible strategies for using this data?
In this course the morning is started with a lecture by a leading biomedical scientist. The topic can be in cancer research, for example, diagnostics or personalised medicine. The presenter will tell us about his/her research and the short term data mining and data integration issues he or she is facing. The lecture is followed by a discussion on possible approaches in solving one or more of these issues. Topics covered will include parsing tabular data, SQL databases, web services and the semantic web. The rest of the day the students will be tasked with finding a solution to a particular problem. Solving such problems can only be done through writing (small) computer programs. This course is suitable for students who take an interest in informatics and biomedical application of informatics. The course builds on the skills acquired in introductionary programming courses; having completed one of these is a hard prerequisite. The introduction to bioinformatics course is not a prerequisite but is highly recommended.
The goal of this course is to outline current data integration challenges in biology and biomedical research and discuss state-of-the-art approaches for tackling these challenges. Students from other disciplines and other universities are invited to attend this course. The topic is suitable for all students in the life sciences dealing with NGS data.